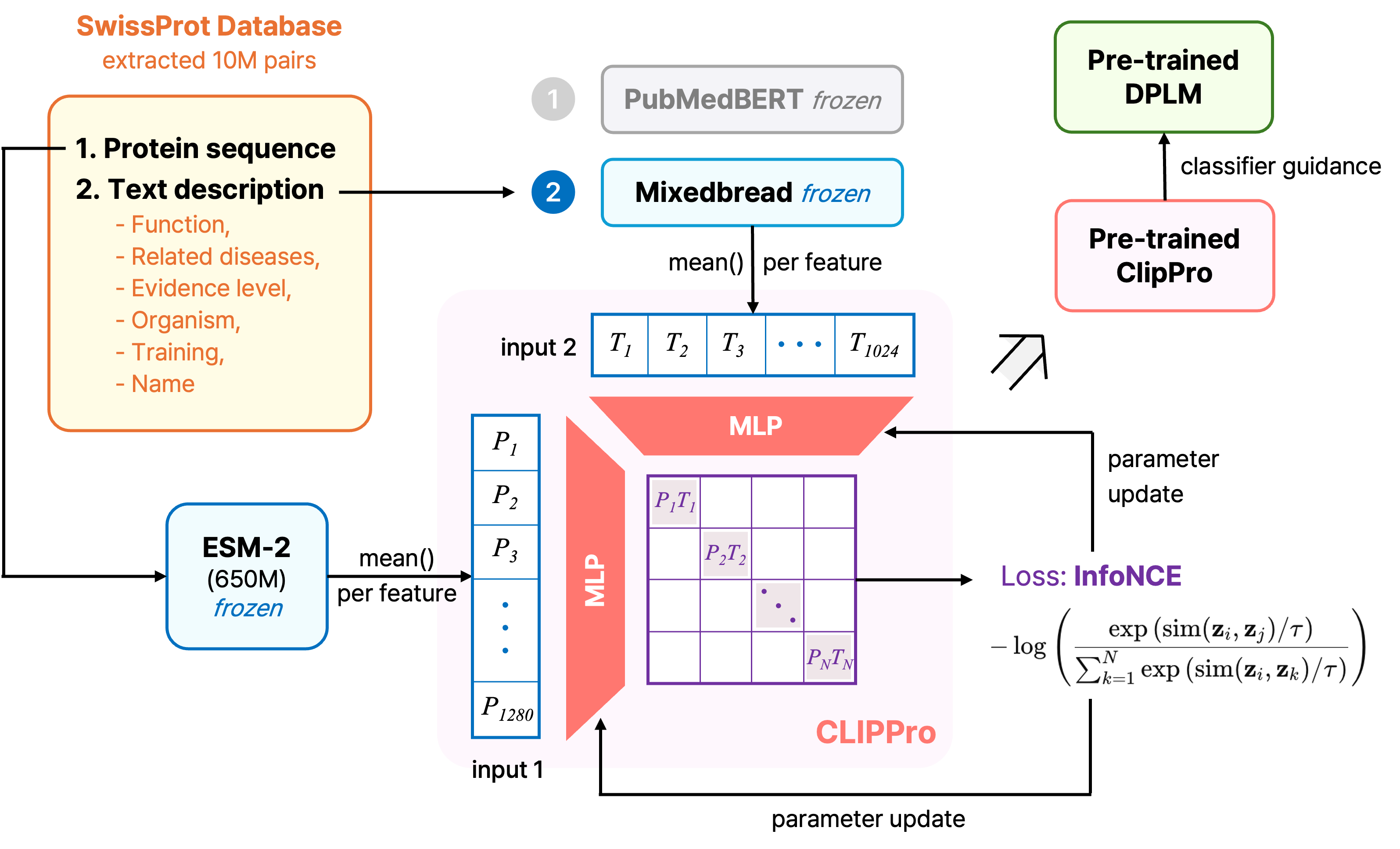
CLIPPro Structure
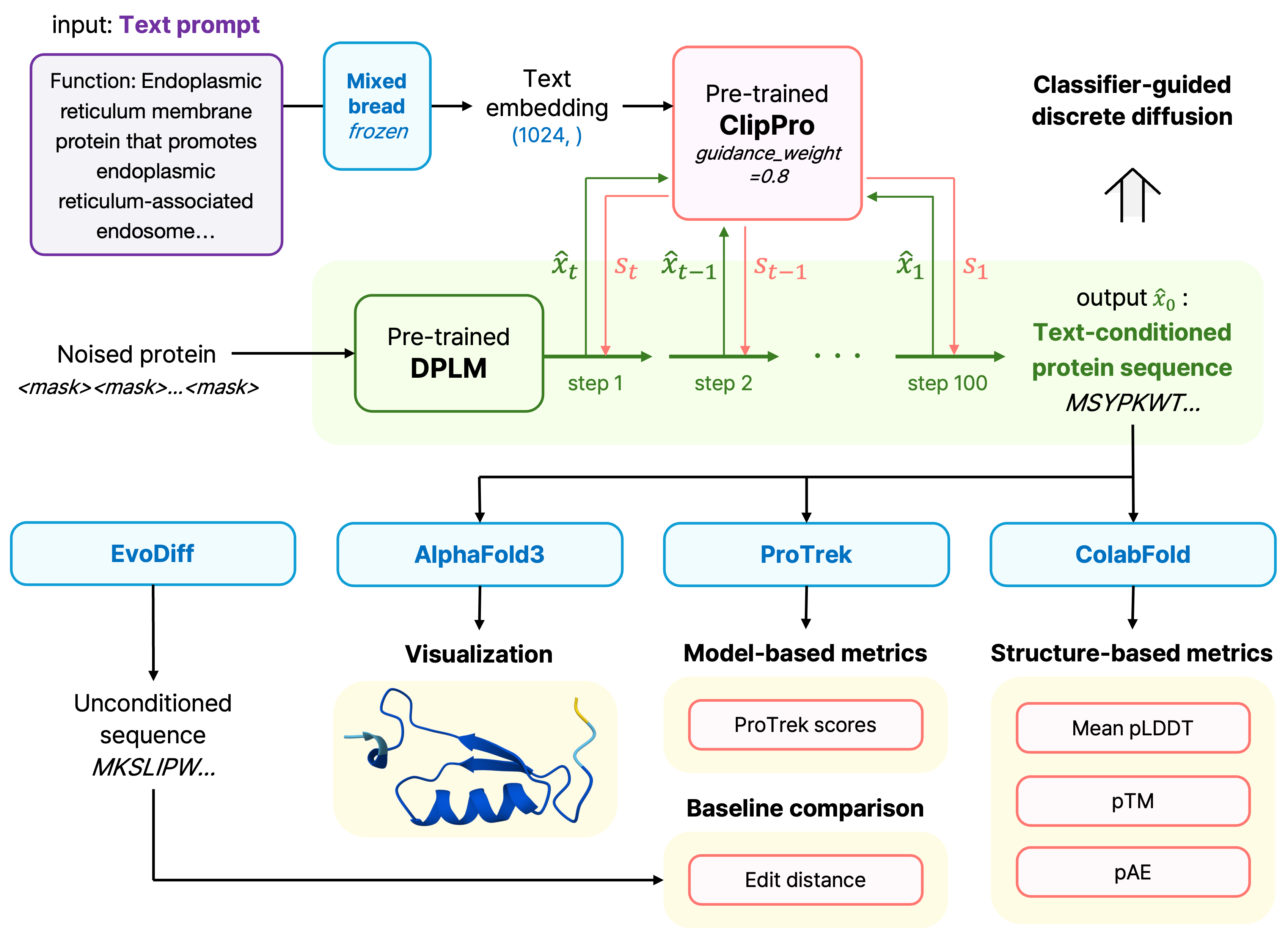
FunctionFold system architecture and generated protein structures, more details available in paper linked here.
We developed FunctionFold, a novel approach for generating functional proteins using natural language input through a classifier-guided diffusion model. We investigated the impact of different natural language encoders (PubMedBERT and Mixedbread) on functional protein generation.
Co-Authors: Yeonseo Kim, Hyojoo Kim, Michaela Harris
Timeline: April 2025
Protein engineering has emerged as a critical tool in biotechnology and drug development, yet current AI-driven approaches primarily focus on structural information or require highly specific technical inputs. While natural language remains the primary medium through which biological function is described, existing protein generation models fail to leverage the nuanced functional descriptions captured in free-form text. Our project addressed this gap by developing FunctionFold, a novel system that generates functional proteins directly from natural language prompts, investigating how different text encoding approaches impact the quality and biological relevance of generated sequences.
By creating a classifier-guided diffusion system, we gained deep insights into the relationship between semantic text representations and protein sequence generation—from contrastive learning principles to the nuances of aligning cross-modal embeddings. This approach allowed us to understand how natural language descriptions can effectively guide protein design toward functionally relevant sequence spaces, while tackling practical challenges in multimodal representation learning and conditional generation.

CLIPPro Structure

FunctionFold system architecture and generated protein structures, more details available in paper linked here.
Our architecture centers on CLIPPro, a custom protein-text alignment model that learns to score how well protein sequences match functional descriptions through contrastive learning. We embedded protein sequences using ESM-2 (650M parameters) and compared two biomedical text encoders: PubMedBERT, trained on biomedical literature, and Mixedbread, optimized for semantic representations. Each embedding passes through modality-specific MLPs projecting into a shared 1024-dimensional latent space, trained using symmetric InfoNCE loss with a learnable temperature parameter.
The trained CLIPPro model then guides the Diffusion Protein Language Model (DPLM) during sequence generation, using cosine similarity scores to compute gradients that steer the diffusion process toward sequences semantically aligned with input prompts. Our system processes natural language descriptions like "binds single-stranded DNA during recombination" or "penetrates cell membranes to deliver molecular cargo," enabling intuitive control over protein generation through biologically meaningful text descriptions rather than technical parameters.
We validated FunctionFold against unguided baselines and EvoDiff across structural and functional metrics using 465,770 protein sequences from SwissProt. Our classifier-guided approach achieved superior functional alignment with 0.2088 Levenshtein similarity compared to 0.2040 for unguided generation, while maintaining competitive structural quality (mean pLDDT of 73.90 vs. EvoDiff's 75.11). Critically, our system demonstrated 16% improvement in ProTrek functional prediction scores (2.30 vs. 1.98), indicating better alignment with intended biological functions.
Ablation studies revealed that semantically meaningful guidance is essential—random text prompts ("apple banana airplane") significantly degraded performance across all metrics, highlighting that appropriate semantic alignment between text and protein representations is crucial for effective generation. Generated proteins exhibited diverse structural characteristics appropriate for their intended functions, from partially disordered algogenic proteins to multi-helical membrane-penetrating structures. While the current implementation focuses on shorter sequences and faces computational overhead during diffusion, these results demonstrate that natural language can effectively guide protein design toward functionally relevant sequence spaces, opening new avenues for democratizing protein engineering through intuitive text-based interfaces.